There Can Be Many Valid Partitions¶
We proclaim that it is in the very spirit of unsupervised learning that, in many cases, there might be many equally valid/plausible/useful ways to partition a dataset (see also [10, 58] for discussion).
Alternative Labellings¶
To take the above postulate into account, we equipped some benchmark sets with alternative labellings. For instance, consider the graves/zigzag_noisy dataset [30]:
import numpy as np
import clustbench
data_url = "https://github.com/gagolews/clustering-data-v1/raw/v1.1.0"
benchmark = clustbench.load_dataset("graves", "zigzag_noisy", url=data_url)
It comes with two different reference partitions:
len(benchmark.labels)
## 2
We depict them below.
import genieclust
for i in range(len(benchmark.labels)):
y_true = benchmark.labels[i]
plt.subplot(1, len(benchmark.labels), i+1)
genieclust.plots.plot_scatter(
benchmark.data, labels=benchmark.labels[i]-1,
axis="equal", title=f"labels{i}; k={benchmark.n_clusters[i]}"
)
plt.show()
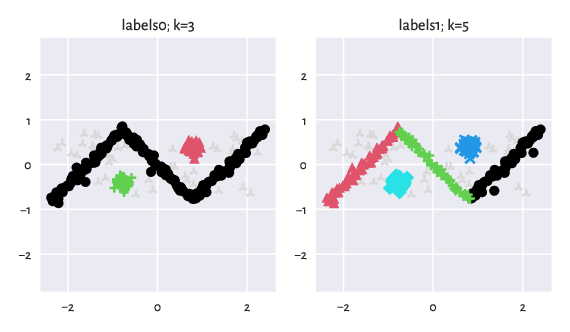
Figure 5: There can be many equally valid partitions.¶
Predicted vs Best-Matching Reference Labelling¶
A clustering algorithm should be rewarded for finding a partition that matches any of the reference ones well. This might require running the method multiple times to find partitions of different cardinalities (unless it is a hierarchical one).
Important
The outputs generated by a single algorithm should be evaluated against all the available reference labellings and the maximal similarity score should be reported.
An example featuring the outputs generated by Genie:
g = genieclust.Genie()
results = clustbench.fit_predict_many(g, benchmark.data, benchmark.n_clusters)
for i, k in enumerate(results):
plt.subplot(1, len(results), i+1)
genieclust.plots.plot_scatter(
benchmark.data, labels=results[k]-1,
axis="equal", title=f"Genie; k={k}"
)
plt.show()
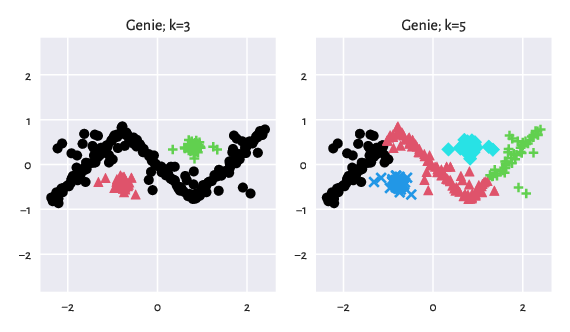
Figure 6: Results generated by Genie.¶
The similarity scores with respect to the consecutive reference label vectors are equal to:
clustbench.get_score(benchmark.labels, results, compute_max=False)
## array([1. , 0.87875611])
And hence the maximal score is:
clustbench.get_score(benchmark.labels, results) # max(the above)
## np.float64(1.0)
Note
Experts (like you) are encouraged to contribute new reference labellings. See also the Colouriser class for a way to generate label vectors interactively (for planar datasets).